Abstract
This study aims to identify genetic causes of familial female infertility characterized by embryonic developmental arrest (EDA) and repeated implantation failure (RIF) with oocyte donation IVF cycle. We used Whole-exome sequencing and Sanger validation to find causative genes in an Iranian consanguineous family that had 3 infertile daughters, 4 fertile daughters, and 2 fertile sons. All patients in this consanguineous family exhibited typical manifestations of unexplained RIF and EDA. Genetic analysis identified a homozygous missense variant (c.G1054C:p.G352R) in exon 13 of the TLE6 gene that cosegregated with the EDA phenotype in an autosomal recessive pattern. Other members of the family, the gene carriers, remain clinically asymptomatic and fertile. Our findings identify a novel nonsynonymous variant, c.G1054C:p.G352R, in the TLE6 gene within a consanguineous Iranian family with autosomal-recessive female infertility and broaden the genetic spectrum of TLE6-associated EDA.
Similar content being viewed by others
Introduction
Infertility is a common health problem defined as failure to conceive after one year of regular unprotected sexual intercourse. It is a notable reproductive health problem that affects more than 10% of people of reproductive age1,2,3. The technique of in vitro fertilization (IVF) is considered as a solution for a large number of infertile couples. Embryonic developmental arrest (EDA) is one of the several factors that can limit the success rate of this technique and is responsible for the high number of embryos that arrest during the first week of in vitro development. Approximately 10–15% of IVF embryos arrest in mitosis at the 2- to 4-cell cleavage stage and show no signs of cell death. Accumulating evidence indicates that this is a common phenomenon in humans. Approximately 10% of all human embryos created by IVF or intracytoplasmic sperm injection become permanently arrested in the early stages of cleavage in culture, and 40% of patients have at least one arrested embryo per treatment cycle4. The permanent arrest of embryos is a non-apoptotic event, since no morphological, no biochemical, and no molecular evidence of apoptosis has been observed before the 8-cell stage in bovine and human embryos5,6,7,8. Since the evaluation of the phenotype associated with EDA in any situation except recurrent IVF failure (RIF) is difficult and accessibility to it in the natural environment is not possible, the genetic cause of EDA in humans is not fully understood. According to previous studies, we can point to chromosomal abnormalities, and single-gene mutations in some genes such as NLRP29, NLRP59, FBXO4310, PADI611, Btg412, CDC2013, MOS14,15, and ACTL7A16 as the genetic causes of EDA. This process is also associated with aberrant regulation of several signaling pathways such as the Wnt and TGFβ signaling pathways17. In the stage of oocyte maturation, oocytes accumulate an essential amount of maternal RNAs and proteins that are used by the early embryo to zygotic genome activation (ZGA)18,19. Maternal factors regulate transcription, both directly and indirectly during ZGA18. Studies in mouse introduced TLE6 as a fundamental maternal effect gene in embryonic preimplantation development20,21. It seems that TLE6 has a pivotal role in symmetric cell division at the 2-cell stage. It stabilized the subcortical maternal complex (SCMC) which controls symmetric cell division in zygotes22,23. TLE6-null embryos have died at the cleavage stage21,22.
WD40-Repeat Proteins (WDR proteins) are one of the largest families encoded by humans24. They play important roles in many fundamental biological processes such as apoptosis25, DNA damage response26, protein degradation27, RNA processing28, transcription regulation29,30, histone modification31, and signal transduction32. WDR proteins often act as scaffolds to engage other molecules, forming protein–protein interactions or functional complexes24. They are classified broadly into 21 classes based on their domain architectures24. Transducin-like enhancer protein (TLE) family belongs to class 7 (TLE_N + WD40)24,33. One such WDR protein, TLE6, belongs to a TLE family that perform numerous critical functions such as controlling and regulating cell cycle progression, gene expression, post-translational modifications, and developmental process33. TLE6 has also been shown as a critical protein required in the embryonic process for female pregnancy. variants in the TLE6 gene at the 19p13.3 locus (OMIM *612399) are responsible for autosomal recessive preimplantation embryonic lethality and EDA. To date, 13 variants have been identified34,35,36,37,38,39,40. The phenotypic spectrum seen in TLE6 mutated patients in these reports suggested sex-associated and genotype–phenotype correlations. Females with bi-allelic variant that result in complete loss of TLE6 active site function exhibit infertility. In contrast, males and females with mono-allelic variant are fertile.
In this study, we characterized the phenotypic spectrum in a consanguineous Iranian family with autosomal recessive EDA due to variant in the TLE6 gene. Here, we report three Iranian infertile sisters due to variant in TLE6. In-depth phenotyping of all members in this consanguineous family pinpoints a clear genotype–phenotype correlation between TLE6 and EDA. We also report a novel variant [NM_001143986(TLE6): c. 1054 G>C (p. G352R)].
Material and methods
Subjects
One Iranian family segregating apparent RIF and EDA was ascertained for this study. Affected individuals underwent clinical examination. Whole blood samples were collected from all family members and genomic DNA was extracted, after obtaining written informed consent. This protocol has been approved by the ethics committee of the Shahid Beheshti and Tehran Universities of Medical Science, Tehran, Iran.
Next-generation sequencing (NGS)
WES (Whole Exome Sequencing) for the family was performed, using the SureSelect XT V6 Human All Exon kit (Agilent Technologies, Santa Clara, CA, USA) following the manufacturer’s protocol. After library quantification and pooling, samples were sequenced on an Illumina NextSeq 500 System (Illumina, San Diego, CA, USA) using the Illumina V3 High Throughput kit. For 3 people in the family (II.11, II.16, II.18) library preparation, sequencing, and bioinformatics analysis were performed using illumina Platform.
Segregation analysis
Segregation analysis was completed by Sanger sequencing on an ABI 3130 Sequencer (Foster City, CA, USA). All sequencing chromatograms were compared to published cDNA sequences for TLE6 (NM_001143986.1), and nucleotide changes were detected using Finch TV Aligner.
Ethics approval and consent to participant
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. Informed consent forms were obtained from all study participants. The study protocol was approved by the ethical committee of Shahid Beheshti University of Medical Sciences and Tehran University of Medical Sciences. All methods were performed in accordance with the relevant guidelines and regulations.
Result
Bioinformatics analysis
Data quality was determined by FASTQC. Paired-end sequences were mapped to the human genome (UCSC hg19) using Burrows-Wheeler Aligner (BWA). Functional variant detection and annotation of genetic variants from high-throughput sequencing data were performed with the GATK (Genome Analysis Toolkit) and ANNOVAR software, respectively. Additionally, variants were filtered with MAF (minor allele frequencies) from the dbSNP, hapmap, Mutation Taster, Intervar, OMIM, Kaviar, 1000 Genomes, gnomAD, ExAC , and the Iranome. First, we filtered variants for quality (depth > 10 and quality score > 30) which was followed by minor allele frequency (MAF) (< 2%). Then, variants were prioritized based on variant-type (missense, nonsense, indel, or splice site), followed by in-silico prediction for conservation (GERP and PhyloP), and predicted deleteriousness (SIFT, PolyPhen2, and the CADD).
For extracting the candidate variants from genomic data of the affected (II.11, II.18) and their healthy sibling (II.16) we consider two hypotheses about infertility and embryonic developmental arrest (EDA) patients.
-
1.
Variants that are associated with dominant diseases:
-
All heterozygous variants are present in the patients and absent in the healthy controls.
-
-
2.
Variants that are associated with recessive diseases:
-
Homozygous variants are present in the patients and heterozygous or absent in the healthy controls.
-
Compound heterozygous variants existing in the patients and missing in the healthy controls.
$$Selected\;Variants=\bigcap_{i=1}^{2}Patients\left(i\right)-\bigcup_{i=1}^{1}Healthy (i)$$
-
By using this pattern, we found 7 variants in 7 genes (WFS1, PLA2G7, IL17F, CHD4, A2ML1, CHRNA4, TLE6). Six of them were placed in the first hypothesis, which we could not find any association for them with infertility and EDA, and one of them (TLE6) was placed in the second hypothesis, we found it in the homozygous format in the affected and heterozygous format in their healthy sister.
Clinical characteristics of the patients
The clinical information of the three patients carrying the biallelic TLE6 variants are listed in Table 1, and their family pedigrees are shown in Fig. 1.
All assessments including karyotypes, menstrual cycle assays, sex hormone levels, and transvaginal sonography revealed no abnormalities. Furthermore, their husbands also showed normal karyotypes and normal semen parameters (sperm morphology, motility, and sperm concentration) (Table 1). This Iranian consanguine family raised seven daughters. Three of them (II-3, II-11, and II-18) had infertility for several years. All the affected individuals underwent several IVF attempts. Two of the affected sisters (II-11 and II-18) even underwent three oocyte donation IVF attempts in the Reproductive Medicine Center.
The II-3 patients (56 years old) had undergone 3 unsuccessful IUI and 3 IVF/ICSI attempts. Totally 18 MII oocytes were retrieved in the three attempts. Only four oocytes were normally fertilized with two pronuclei (PN) zygotes, while the others were abnormally fertilized with 0PN or degradation on day 1. After cultivation, most of her embryos were arrested at the early stages with heavy fragmentation, and only two poor-quality embryos were available for transfer. Although the patient underwent 2 embryo transfer cycles, she failed to obtain a successful pregnancy. Her spouse got married and now has 2 children.
The other affected sister (II-11, 37 years old) underwent 1 unsuccessful IUI and 3 IVF/ICSI attempts with her oocytes and 3 IVF/ICSI attempts with donated oocytes. A total of 14 MII oocytes were retrieved in the 3 attempts. Some of her oocytes were abnormally fertilized with 0PN; 7 of them showed normal fertilization with 2PN zygotes on day 1. All of her embryos were arrested at the early stages having heavy fragmentation. Finally, after hysteroscopy and IVF with oocyte donation she got pregnant. In 3 IVF attempts with donated oocyte, 2 first attempts were failed and finally, in the last one, she got pregnant. Before the 3rd attempt, diagnostic hysteroscopy was performed on her and everything was normal. Her spouse's information is available on the Table 1.
The proband (II-18, 32 years old) underwent 1 unsuccessful IUI, 3 IVF/ICSI attempts with her oocytes, and 3 IVF/ICSI attempts with donated oocytes. A total of 19 MII oocytes were retrieved in the three attempts. Only 2 oocytes were normally fertilized with two pronuclei (PN) zygotes, while the others were abnormally fertilized with 0PN or degradation on day 1. After cultivation, all of her embryos were arrested at the early stages with heavy fragmentation. Totally, in 3 IVF attempts with donated oocyte, 2 first attempts were failed and finally, in the last one, she got pregnant. Before the 3rd attempt, diagnostic hysteroscopy was performed on her and everything was normal. Her spouse's information is available in the Table 1 and is completely normal.
Identification of novel variant in TLE6
All patients in this consanguineous family exhibited typical manifestations of unexplained RIF and EDA. Genetic analysis identified a homozygous missense variant (c.G1054C:p.G352R) in exon 13 of the TLE6 gene that cosegregated with the EDA phenotype in an autosomal recessive pattern (NCBI, ClinVar accession number is SCV002525878.). Other members of the family, gene carriers, remain clinically asymptomatic and fertile.
The homozygous variant [NM_001143986(TLE6): c.1054G>C (p. G352R)] was identified in the proband and all of her affected sisters. This variant was verified by Sanger sequencing. Both of the parents, four fertile elder sisters, and one elder brother were heterozygous carriers, indicating a recessive inheritance pattern (Fig. 1). This variant has not been reported in ClinVar, HGMD, gnomAD, Iranome, ExAC, dbSNP, and 1000G. Based on this situation this is a novel variant (Tables 2 and 3).
Results of conservative and in silico analysis
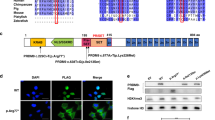
The amino acids at position p.G352 of TLE6 were highly conserved in 141 different taxonomies, suggesting that the variant was likely pathogenic. The location of the TLE6 variant and the conservation analysis among different species are shown in Fig. 2. The variant identified in this study (c.G1054C:p.G352R) resides in the buried residue of the WDR2 domain and seems to play a central role in the enzyme activity (Fig. 3). According to the three-dimensional (3D) structure of TLE6, the nonsynonymous variant caused a replacement of glycine with arginine at position 352, leading to the production of 30 new binding sites and deactivation of all of its active sites and 20 native binding sites compared with the Wild type (Fig. 3).
The locations and conservation of mutated residues in TLE6 (TLE6:NM_001143986:exon13:c.G1054C:p.G352R). (A) The position of the variant is indicated in the genomic structure and protein structure of TLE6. (B) Conservation of mutated amino acids in TLE6 among 141 different taxonomies. The residue G352 is highly conserved across species.
By using I-Taster, we found that TLE6 has 3 active site residues and 33 binding site residues. G352R variant gives rise to loss of all of its active site residues and 20 native binding site residues and achievement of 30 new binding sites, as a result, this variant caused TLE6 to have no active site and 43 binding sites (For more detail refer to attachments).
Phenotypic spectrum of the patients with TLE6 variant p.G352R
We used light microscopy to observe the development and morphology of the embryos from family members II-5 and II-8 for 5 consecutive days in their last ICSI attempt. Five of the embryos on day 3 were arrested, whereas the others had a high percentage of fragmentation, and all of them failed to form blastocysts (Fig. 4).
The morphology of the embryo is derived from the control and the proband, respectively.
Discussion
Many infertile couples have experienced recurrent IVF/ICSI failed attempts. In some of them, the obtained oocytes and ovulatory status look normal, but zygote formation and embryonic development are severely impaired. This study aims to identify genetic causes of familial female infertility characterized by EDA and RIF. We used whole-exome sequencing and Sanger validation to find causative genes in an Iranian consanguineous family that has 9 children, 3 infertile daughters, 4 fertile daughters, and 2 fertile sons. All patients in this consanguineous family exhibited typical manifestations of unexplained RIF and EDA. Genetic analysis identified a novel homozygous missense variant (c.G1054C:p.G352R) in exon 13 of the TLE6 gene that segregated with the EDA phenotype in an autosomal recessive pattern. Other members of the family, gene carriers, remain clinically asymptomatic and fertile.
TLE (Transducin-like enhancer protein) family is a conserved family of corepressor proteins, which cannot bind DNA directly but repress transcription by interacting with partner proteins33,42. They repress gene expression through different mechanisms43. Two evolutionarily conserved domains are found in the TLE family: the carboxy-terminal WD40 repeat domain and an amino-terminal (TLE N-terminal), also known as the Q-rich domain. It seems that the Q-rich domain is important for oligomerization of TLE and binding to specific transcription factors and interactions with most transcription factors are mediated by the WD40 repeat domain33,43,44.
WD40-Repeat Proteins (WDR proteins) are one of the largest families encoded by humans7. They play important roles in many fundamental biological processes such as apoptosis8, DNA damage response17, protein degradation18, RNA processing18, transcription regulation20,21, histone modification22, and signal transduction23. WDR proteins often act as scaffolds to engage other molecules, forming protein–protein interactions or functional complexes7. They are classified broadly into 21 classes based on their domain architectures7. Transducin-like enhancer protein (TLE) family belongs to Class 7 (TLE_N + WD40)7,24.
There are four full-length TLE genes in humans named TLE1-4 and three truncated forms, TLE5-7. The truncated TLE isoforms are thought to inhibit the function of the full-length TLEs (TLE1-4)33. TLE6 is located on chromosome 19 at 19p13.3. It contains 17 exons that span more than 17 kb of genomic DNA and it encodes the Transducing-like enhancer protein 6 (Q9H808- TLE6- Human) with 572 amino acids. Expressing in different tissue such as the placenta, thyroid, lung, and testis. It has 7 WDR domains.
To date, 13 variants have been identified in TLE6—associated with EDA characterized by female fertility34,35,36,37,38,39,40. 64% of them were located in the WDR domain. This paper adds the 14th variant which is located in the WDR domain. The variant identified in this study (c.G1054C:p.G352R) leads to the replacement of Gly 352 with Arg residing in the buried residue of the WDR2 domain which plays a central role in enzyme activity. It is speculated that native TLE6 has 3 Active site residues (156,125 and 145) and 33 ligand binding sites. This variant leads to missing all active sites and 20 native binding sites and also it leads to gaining 30 new binding sites.
By considering its roles in signaling pathways (Notch and Wnt)45,46 and some biological functions such as endoplasmic reticulum localization, spindle localization, mitochondrion localization, regulation of cell division, and regulation of transcription by RNA polymerase II, it would be clear that loss of its functions may lead to infertility.
In summary, this study extended the spectrum of genetic causes of familial female infertility characterized by EDA by reporting a novel variant in TLE6. Our result suggests oocyte donation as the best ART method for patients with biallelic TLE6 variants right now.
Data availability
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request. The identified variant in this study has been deposited into NCBI, ClinVar (accession number: SCV002525878) and will be publicly available per NCBI "Hold until publish" policy.
Change history
13 December 2022
A Correction to this paper has been published: https://doi.org/10.1038/s41598-022-26129-7
References
VC, R. R., Jogi, P., Bolem, O., Gujju, E. & Sanaboina, A. A review on risk factors, staging and survival rates of endometrial cancer in both black and white women in infertility patients in USA. World J. Curr. Med. Pharm. Res. 2, 152–156 (2020).
Morshed-Behbahani, B., Lamyian, M., Joulaei, H., Rashidi, B. H. & Montazeri, A. Infertility policy analysis: A comparative study of selected lower middle-middle-and high-income countries. Glob. Health 16, 1–9 (2020).
Moghadam, A. D., Delpisheh, A. & Sayehmiri, K. The trend of infertility in Iran, an original review and meta-analysis. Nursing Practice Today 1, 46–52 (2014).
Betts, D. & Madan, P. Permanent embryo arrest: Molecular and cellular concepts. MHR Basic Sci. Reproduct. Med. 14, 445–453 (2008).
Byrne, A., Southgate, J., Brison, D. & Leese, H. Analysis of apoptosis in the preimplantation bovine embryo using TUNEL. Reproduction 117, 97–105 (1999).
Hardy, K. Apoptosis in the human embryo. Rev. Reprod. 4, 125–134 (1999).
Matwee, C., Betts, D. H. & King, W. A. Apoptosis in the early bovine embryo. Zygote 8, 57–68 (2000).
Gjørret, J. O. et al. Chronology of apoptosis in bovine embryos produced in vivo and in vitro. Biol. Reprod. 69, 1193–1200 (2003).
Mu, J. et al. Mutations in NLRP2 and NLRP5 cause female infertility characterised by early embryonic arrest. J. Med. Genet. 56, 471–480 (2019).
Wang, W. et al. FBXO43 variants in patients with female infertility characterized by early embryonic arrest. Hum. Reprod. 36, 2392–2402 (2021).
Wang, X. et al. A novel homozygous mutation in the PADI6 gene causes early embryo arrest. Reprod. Health 19, 1–11 (2022).
Yu, C. et al. BTG4 is a meiotic cell cycle–coupled maternal-zygotic-transition licensing factor in oocytes. Nat. Struct. Mol. Biol. 23, 387–394 (2016).
Zhao, L. et al. Biallelic mutations in CDC20 cause female infertility characterized by abnormalities in oocyte maturation and early embryonic development. Protein Cell 11, 921–927 (2020).
Zhang, Y. L. et al. Biallelic mutations in MOS cause female infertility characterized by human early embryonic arrest and fragmentation. EMBO Mol. Med. 13, e14887 (2021).
Zeng, Y. et al. Bi-allelic mutations in MOS cause female infertility characterized by preimplantation embryonic arrest. Hum. Reprod. 37, 612–620 (2022).
Xin, A. et al. Disruption in ACTL7A causes acrosomal ultrastructural defects in human and mouse sperm as a novel male factor inducing early embryonic arrest. Sci. Adv. 6, eaaz4796 (2020).
Mohebi, M. & Ghafouri-Fard, S. Embryo developmental arrest: Review of genetic factors and pathways. Gene Rep. 17, 100479 (2019).
Wu, D. & Dean, J. Maternal factors regulating preimplantation development in mice. Curr. Top. Dev. Biol. 140, 317–340 (2020).
Luong, X. G. & Conti, M. Human Reproductive and Prenatal Genetics 193–220 (Elsevier, 2019).
Feng, M., Bai, Y., Chen, Y. & Wang, K. Knockout of the Transducin-like enhancer of split 6 gene affects the proliferation and cell cycle process of mouse spermatogonia. Int. J. Mol. Sci. 21, 5827 (2020).
Yu, X.-J. et al. The subcortical maternal complex controls symmetric division of mouse zygotes by regulating F-actin dynamics. Nat. Commun. 5, 1–12 (2014).
Zhu, K. et al. Identification of a human subcortical maternal complex. MHR Basic Sci. Reproduct. Med. 21, 320–329 (2015).
Bebbere, D., Albertini, D. F., Coticchio, G., Borini, A. & Ledda, S. The subcortical maternal complex: Emerging roles and novel perspectives. Mol. Hum. Reproduct. 27, gaab043 (2021).
Zou, X.-D. et al. Genome-wide analysis of WD40 protein family in human. Sci. Rep. 6, 1–11 (2016).
Zou, H., Henzel, W. J., Liu, X., Lutschg, A. & Wang, X. Apaf-1, a human protein homologous to C. elegans CED-4, participates in cytochrome c–dependent activation of caspase-3. Cell 90, 405–413 (1997).
Wakasugi, M. et al. DDB accumulates at DNA damage sites immediately after UV irradiation and directly stimulates nucleotide excision repair. J. Biol. Chem. 277, 1637–1640 (2002).
Higa, L. A. et al. CUL4–DDB1 ubiquitin ligase interacts with multiple WD40-repeat proteins and regulates histone methylation. Nat. Cell Biol. 8, 1277–1283 (2006).
Yan, C. et al. Structure of a yeast spliceosome at 3.6-angstrom resolution. Science 349, 1182–1191 (2015).
Jennings, B. H. et al. Molecular recognition of transcriptional repressor motifs by the WD domain of the Groucho/TLE corepressor. Mol. Cell 22, 645–655 (2006).
Znaidi, S., Pelletier, B., Mukai, Y. & Labbé, S. The Schizosaccharomyces pombe corepressor Tup11 interacts with the iron-responsive transcription factor Fep1. J. Biol. Chem. 279, 9462–9474 (2004).
Ruthenburg, A. J. et al. Histone H3 recognition and presentation by the WDR5 module of the MLL1 complex. Nat. Struct. Mol. Biol. 13, 704–712 (2006).
Gaudet, R., Bohm, A. & Sigler, P. B. Crystal structure at 2.4 Å resolution of the complex of transducin βγ and its regulator, phosducin. Cell 87, 577–588 (1996).
Agarwal, M., Kumar, P. & Mathew, S. J. The Groucho/Transducin-like enhancer of split protein family in animal development. IUBMB Life 67, 472–481 (2015).
Mao, B. et al. A novel TLE6 mutation, c. 541 + 1G> A, identified using whole-exome sequencing in a Chinese family with female infertility. Mol. Genet. Genom. Med. 9, e1743 (2021).
Liu, J. et al. Two novel mutations in PADI6 and TLE6 genes cause female infertility due to arrest in embryonic development. J. Assist. Reprod. Genet. 38, 1551–1559 (2021).
Wang, L.-Q. et al. LncRNA-Fendrr protects against the ubiquitination and degradation of NLRC4 protein through HERC2 to regulate the pyroptosis of microglia. Mol. Med. 27, 39. https://doi.org/10.1186/s10020-021-00299-y (2021).
Zheng, W. et al. Expanding the genetic and phenotypic spectrum of the subcortical maternal complex genes in recurrent preimplantation embryonic arrest. Clin. Genet. 99, 286–291 (2021).
Lin, J. et al. Expanding the genetic and phenotypic spectrum of female infertility caused by TLE6 mutations. J. Assist. Reprod. Genet. 37, 437–442 (2020).
Wang, X. et al. Novel mutations in genes encoding subcortical maternal complex proteins may cause human embryonic developmental arrest. Reprod. Biomed. Online 36, 698–704 (2018).
Alazami, A. M. et al. TLE6 mutation causes the earliest known human embryonic lethality. Genome Biol. 16, 1–8 (2015).
Zhang, M. et al. Identification of novel biallelic TLE6 variants in female infertility with preimplantation embryonic lethality. Front. Genet. 894 (2021).
Beagle, B. & Johnson, G. V. AES/GRG5: More than just a dominant-negative TLE/GRG family member. Dev. Dyn. 239, 2795–2805 (2010).
Turki-Judeh, W. & Courey, A. J. Groucho: A corepressor with instructive roles in development. Curr. Top. Dev. Biol. 98, 65–96 (2012).
Jennings, B. H. & Ish-Horowicz, D. The Groucho/TLE/Grg family of transcriptional co-repressors. Genome Biol. 9, 1–7 (2008).
Salazar, J. L. & Yamamoto, S. Integration of Drosophila and human genetics to understand notch signaling related diseases. Mol. Mech. Notch Signaling. 1066, 141–185 (2018).
Tribulo, P. et al. WNT regulation of embryonic development likely involves pathways independent of nuclear CTNNB1. Reproduction 153, 405–419 (2017).
Acknowledgements
This study was financially supported by both Shahid Beheshti University of Medical Sciences and Tehran University of Medical Sciences.
Author information
Authors and Affiliations
Contributions
Conceptualization: S.G.F., M.H.M., M.A., M.M., K.B. Data curation: M.A., A.G. Formal analysis: M.A., M.H.M., K.B. Methodology: M.A., A.G. Visualization: M.A., M.H.M., K.B. Writing original draft: M.A., M.M. Writing review and editing: M.A., M.M., and S.G.F. Supervision: S.G.F., M.H.M.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
The original online version of this Article was revised: The original version of this Article contained an error in the spelling of the author Mohammad Hossein Modarressi, which was incorrectly given as Mohammad-Hossein Modarresi.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Akbari, M., Mohebi, M., Berjis, K. et al. A novel variant in TLE6 is associated with embryonic developmental arrest (EDA) in familial female infertility. Sci Rep 12, 17664 (2022). https://doi.org/10.1038/s41598-022-22687-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-022-22687-y
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.